Microscopy encompasses a variety of methods for obtaining research data with high spatial and temporal resolution and is a methodological pillar throughout the life sciences and medicine. Various methods allow image data to be generated with the aid of light or electron beams. These data are visualized as microscopy images, and quantitative information about the investigated object can be obtained using image analysis. Specific sample preparation protocols allow the visualization and measurement of specific structures. Example methods include confocal laser microscopy with fluorescent dyes, as well as reflected light and transmitted light wide-field imaging using diverse staining methods. Not only do procedures such as high-throughput microscopy or special techniques such as light sheet microscopy deliver very large amounts of data. The individual image files also often have a complex structure (4-, 5-, or multi-dimensional) and can reach enormous individual file sizes (GB to TB). At the same time, the file formats and metadata structures of the various microscope vendors are to date not well standardized.
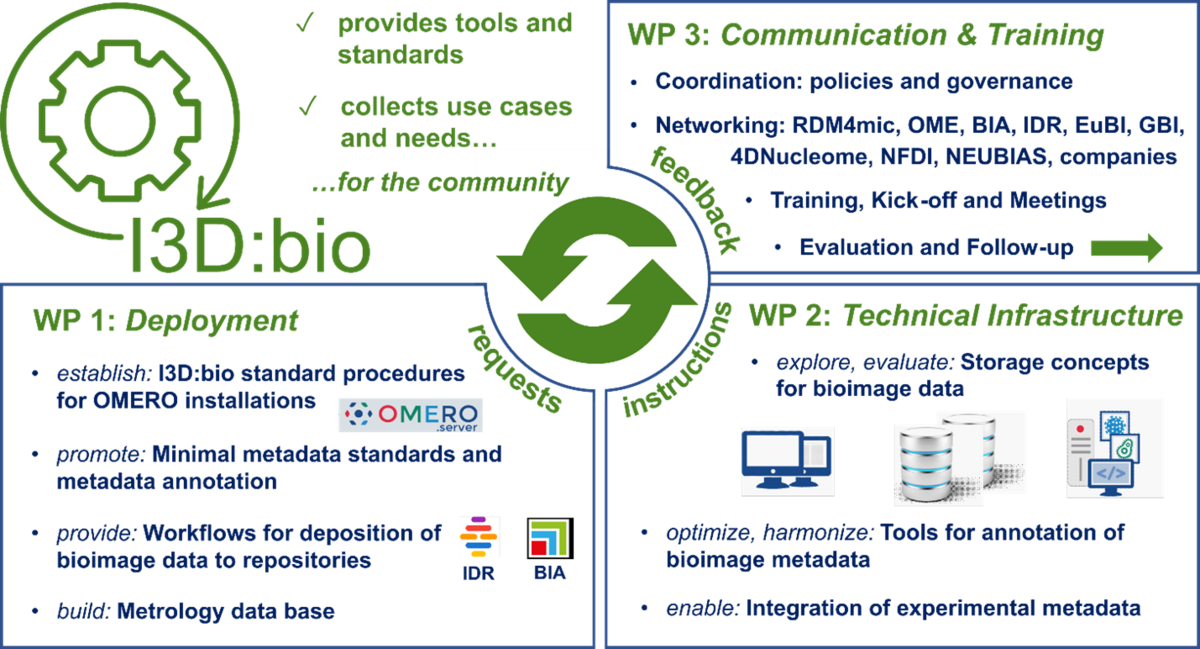
Therefore, research data management in the field of microscopy poses challenges to researchers. The Information Infrastructure for BioImage Data (I3D:bio) project, funded by the German Research Foundation (DFG), addresses solutions for research data management and sharing in bioimaging. One focus of the project is the supported implementation of the open software OMERO (OME Remote Objects), which was developed specifically for image data management by the Open Microscopy Environment Consortium. The I3D:bio project develops target-group-specific installation guides and supports partner institutions at distributed locations in Germany in the implementation of OMERO. In particular, the light microscopy core facilities shall be enabled to support their users in image data management using OMERO. In addition, the project partners are developing new solutions for the interoperability of OMERO with electronic lab notebooks and recommendations for standardized metadata annotation in Tags and Key-Value pairs in OMERO. The immediate benefits for OMERO users are highlighted, and training materials for using OMERO are developed. Improved image data management starting at data acquisition is also a means to ease the deposition of published datasets in image data repositories like the BioImage Archive or the Image Data Resource. In another subproject, better comparability of quality controls on microscopes prior to image acquisition is intended. For this purpose, a repository based on OMERO will be created (metrology database). Better comparability of protocols in equipment management will increase the quality of the resulting image data and provide a basis for comparability of publicly shared research data in bioimaging. The overall project goal is to facilitate the implementation of the FAIR principles (findability, accessibility, interoperability, and reusability) for biological image data in everyday research practice.
(copy 2)
Services
- Help Desk for the implementation of OMERO at universities and research institutes
- Metrology database (under development)
- Knowledge-Hub for Bioimaging-related research data management topics
- Training material for the use of OMERO
Project Partners
- Heinrich Heine University Düsseldorf, Center for Advanced Imaging
- DKFZ German Cancer Research Center, Dep. Enabling Technology
- University of Osnabrück, Integrated Bioimaging Facility Osnabrück, CellNanOs
- University of Freiburg, Life Imaging Center
- Supported by German BioImaging – Society for Microscopy and Image Analysis
- I3D:bio collaborates closely with the NFDI consortium NFDI4BIOIMAGE
Project duration
01.01.2022 – 31.12.2024
Project Funding
- German Research Foundation (Deutsche Forschungsgemeinschaft, DFG), grant number 462231789
Publications
- Kunis, Susanne, & Dohle, Julia. (2022). Structuring of Data and Metadata in Bioimaging: Concepts and technical Solutions in the Context of Linked Data. Zenodo. https://doi.org/10.5281/zenodo.7018750
- I3D:bio's OMERO Training Videos 2023, https://www.youtube.com/playlist?list=PL2k-L-zWPoR7SHjG1HhDIwLZj0MB_stlU, based on: Schmidt, C., Bortolomeazzi, M., Boissonnet, T., Fortmann-Grote, C., Dohle, J., Zentis, P., Kandpal, N., Kunis, S., Zobel, T., Weidtkamp-Peters, S., & Ferrando-May, E. (2023). I3D:bio's OMERO training material: Re-usable, adjustable, multi-purpose slides for local user training. Zenodo. https://doi.org/10.5281/zenodo.832358
- Schmidt C, Hanne J, Moore J et al. Research data management for bioimaging: the 2021 NFDI4BIOIMAGE community survey. F1000Research 2022, 11:638 (https://doi.org/10.12688/f1000research.121714.2)
- Schmidt, C., Bortolomeazzi, M., Boissonnet, T., Fortmann-Grote, C., Dohle, J., Zentis, P., Kandpal, N., Kunis, S., Zobel, T., Weidtkamp-Peters, S., & Ferrando-May, E. (2023). I3D:bio's OMERO training material: Re-usable, adjustable, multi-purpose slides for local user training. Zenodo. https://doi.org/10.5281/zenodo.8323588
- Schmidt, Christian et al.: Bringing FAIR Bioimage Data Management into Practice: the Information Infrastructure for BioImage Data (I3D:bio) Project – bottom-up Community Support for Microscopy Data Sharing and Preservation., in Heuveline, Vincent, Bisheh, Nina und Kling, Philipp (Hrsg.): E-Science-Tage 2023: Empower Your Research – Preserve Your Data, Heidelberg: heiBOOKS, 2023, S. 289–294. https://doi.org/10.11588/heibooks.1288.c18090
Further Information
- Website and Knowledge Hub
- Regular meetings of the Research Data Management for Microscopy (RDM4mic) group, from which the project arose
- German BioImaging
Contact
Get in touch via our website: https://gerbi-gmb.de/i3dbio/i3dbio-help-contact/
e-mail: i3dbio@gerbi-gmb.de